R
R provides a set of base functions, which can
be extended by using contributed packages (to be installed additionally).
Say, we needed such a contrib-package and therefore download
and install the MASS library from
Venable and Ripley. MASS provides many statistical functions and also
numerous sample datasets:
#don't get confused: the code package is called VR, the library MASS:
install.packages("VR")
library(MASS)
To get R command help, you can always add a ? to the command:
?library #or open the browser: help.start()
Additionally there is the FAQ).
To get out of this (if already feeling sleepy):
q()
R is an object oriented data analysis language. The assignment operator is used to store results:
x <- 1+1
To get the result, just enter the name of the object:
x
A simple example is to plot a sine curve. First we have to define the interval and number of data points to support the curve. Note: By redefining x we overwrite the old object:
x <- seq(-2*pi, 2*pi, len = 100) x str(x) summary(x)
Then we can plot it (the type parameter specifies the line type):
matplot(x, sin(x), type="l")
To get an idea about the various "matplot()" options, run:
?matplot
You can see some "matplot()" examples by running:
example(matplot)
R comes along with a large number of published datasets. To get a list about currently accessible datasets, enter:
data()
To get a description of a specific dataset, enter (e.g. volcano dataset):
?volcano
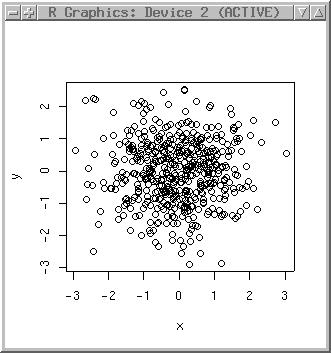
Before using such data, we generate a set of coordinate pairs (500 points) normally distributed (just to get some experience with R):
x <- rnorm(500) y <- rnorm(500)
Plot them:
plot(x,y)
Look at the histogram:
library(MASS) truehist(x) truehist(y, nbin=50)
The nbins parameter allows to define the number of bins (those bars you can see there) in the histogram.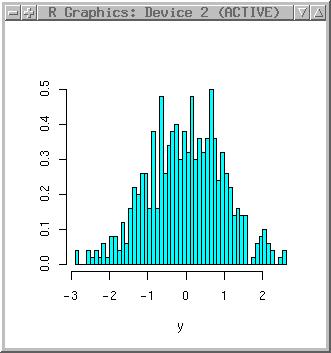
Let's add some z-values. To get numbers larger that 1 we multiply the values generated by rnorm:
z <- 1000 * rnorm(500)
To proceed, we need to link these three variables. They can be bounded into a "data frame":
dataset <- data.frame(x,y,z)
Now we have these variables twice, once individual, once in a dataframe. To see the objects currently available, use:
ls()
You can look at an object by entering its name. Here you will receive a long list of numbers with four columns (number of element, x, y, z):
dataset
Get a summary of it:
summary(dataset)
To look at the data structure (you will need to do this regularly), use the str() function:
str(dataset)
As we have a dataframe now, the access to a variable is different:
mean(x)
mean(dataset$x)
Hopefully you get the same results. Generally you could remove the individual variables if you will continue to work with the dataframe:
rm(x,y,z) ls()
library(akima) surface <- interp(dataset$x, dataset$y, dataset$z) contour(surface)
We want to see a colored surface with legend:
filled.contour(surface)
... and a perspective view:
persp(surface, col="green", expand=0.3)
New developments (2003) such as iPlots and RGL will improve the visualization capabilities of R. Also xyplot() of the lattice library is very powerful.
Finally we plot a color filled sine curve:
# set "frame": plot(c(-pi, 2*pi), c(-1,1), type="n", ylab="sin(x)", xlab="x") x1 <- seq(-pi, 0, length=101) x2 <- seq(0, 2*pi, length=101) polygon(c(0, -pi, x1), c(0, 0, sin(x1)), col="red") polygon(c(2*pi, 0, x2), c(0, 0, sin(x2)), col="green") # print plot into EPS file: dev.copy2eps()
x <- dataset$x
To export this single variable, use:
write(x, file="~/testfile.asc")
To export a dataframe, use:
write.table(dataset, file="~/testfile2.asc")
You probably want to use a column separator different from white space:
write.table(dataset, file="~/testfile3.asc", sep=":")
Read the table back into R:
dataset.new <- read.table("~/testfile.asc", header=T, sep=":")
Remove the x, y and z variable from memory of R (we still have the dataframe):
rm(x)
List available objects:
ls()
Now we want to get a summary of the dataframe variables (Min, 1st. Quartile, Median, Mean, 3rd Qu., Max, NA's):
summary(dataset)
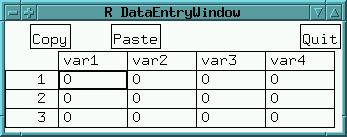
If you want to enter data tables from keyboard, R provides the function data.entry(). To enter a 3x4 matrix with value 0 pre-defining the fields, enter:
x <- matrix(0,3,4) data.entry(x)
postscript("test.ps")
plot(x)
dev.off()
xfig("test.fig")
plot(x)
dev.off()
dev.copy2eps()
# may not work on all systems:
install.packages("rgl", dependencies=TRUE)
# does not neccesarily need 'rgl':
install.packages("Rcmdr", dependencies=TRUE)
library(Rcmdr)
To re-lauch it, run:
Commander()
End of basic stuff.